EagleEye
Navigation
Our Sponsors
Tool: Gene Set Clustering based on Functional annotation (GeneSCF)
-----------
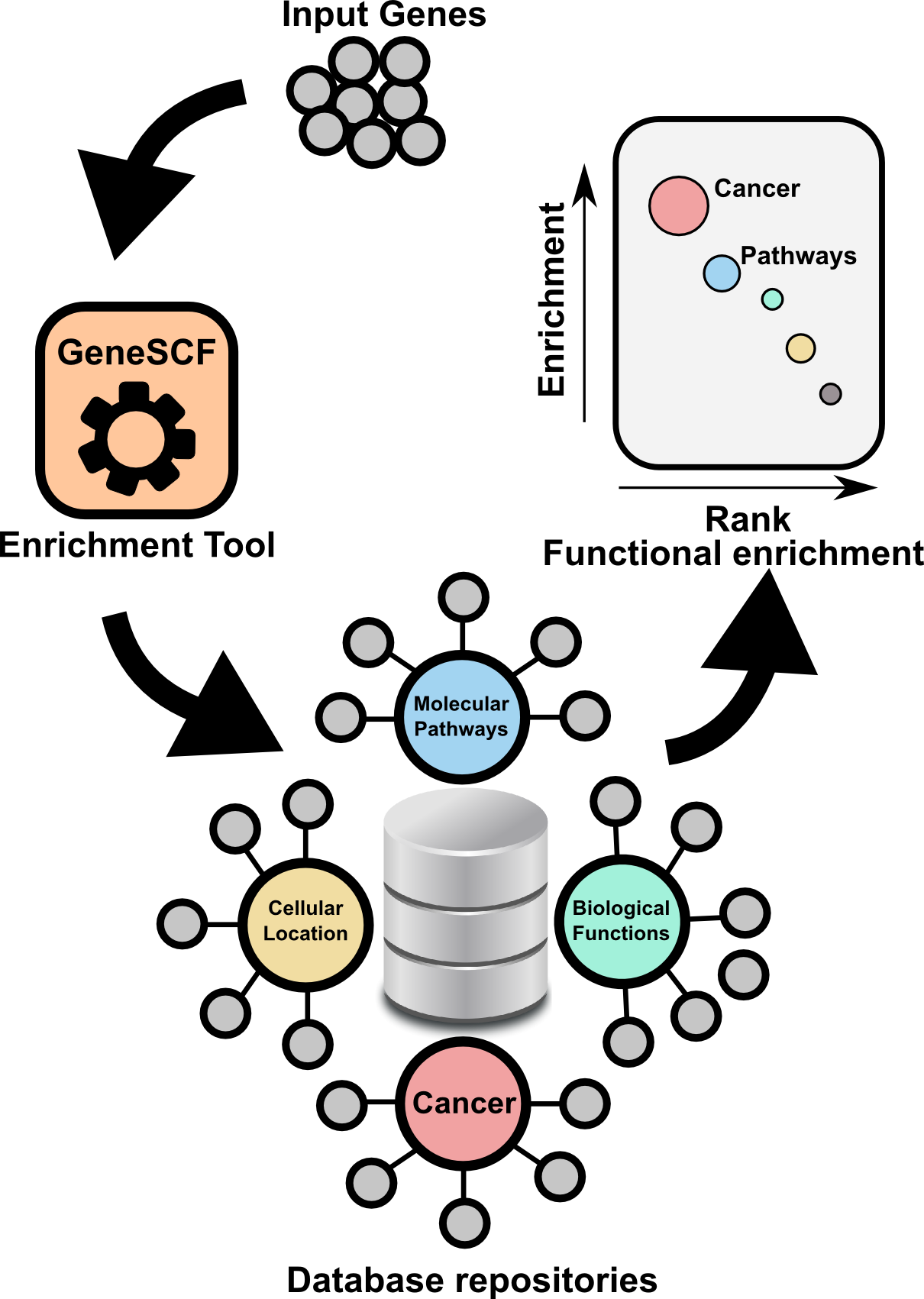
Gene Set Clustering based on Functional annotation
GeneSCF serves as command line tool for clustering the list of genes given by the users based on functional annotation (Gene Ontology, KEGG, REACTOME and NCG 4.0). It requires gene list in the form of Entrez Gene ID (UIDs) or Official gene symbols as a input. GeneSCF supports more organisms from V1.1. Examples to download database as simple text file using GeneSCF "prepare_database" module, 1) https://www.biostars.org/p/197414/#197416 , 2) https://www.biostars.org/p/191532/#191540
The advantage of using GeneSCF over other enrichment tools is that, it performs enrichment analysis in real-time (v1.1 and above) by accessing source databases. With command-line versions of tools, as you know you can run multiple gene list simultaneously.
------------
Home page: http://genescf.kandurilab.org/ |
Requirement:
GeneSCF only works on Linux system, it has been successfully tested on Ubuntu, Mint and Cent OS. Other distributions of Linux might work as well.
Documentation:
http://genescf.kandurilab.org/documentation.php
Report issues on Biostars or GitHub Project page
----------
- Published: GeneSCF: a real-time based functional enrichment tool with support for multiple organisms
2775 days ago
EagleEyeNews - Pathway Analysis
3492 days ago
Rahul Agarwaltop level page - KOALA: KEGG's internal annotation tool for K number assignment of KEGG GENES using SSEARCH...
1961 days ago
Abhimanyu Singhbookmark - ClueGO
2884 days ago
Jitbookmark - WEGO : simple but useful tool for visualizing, comparing and plotting GO (Gene Ontology) annotation...
1474 days ago
BioStarbookmark - Understanding GO analysis
443 days ago
Neelam Jhabookmark - R package for visualising GO enrichment
3930 days ago
JitNews - BioDBnet
2884 days ago
Jitbookmark

