Anjana
Bioinformatics enthusiast
Our Sponsors
Murasaki
http://murasaki.dna.bio.keio.ac.jp/wiki/index.php?Murasaki
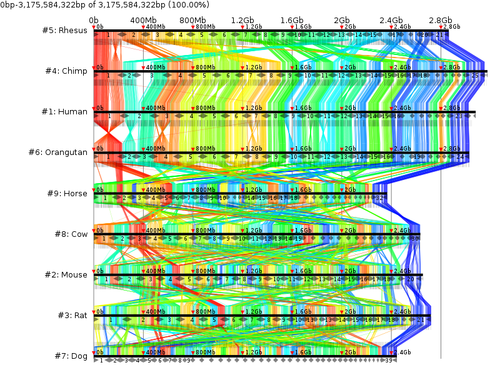
Murasaki is an anchor alignment program that is
- exteremely fast (17 CPU hours for whole Human x Mouse genome (with 40 nodes: 35 wall minutes), or 8 mammals in 21 CPU hours (42 wall minutes))
- scalable (Arbitrarily parallelizable across multiple nodes using MPI)
- memory efficient. (Even a single node with 16GB of ram can handle over 1Gbp of sequence)
- unlimited by pattern length or selection
- repeat tolerant
Related items
- GenomeRing: alignment visualization based on SuperGenome coordinates
3334 days ago
Neelbookmark - Strudel
3444 days ago
Anjanabookmark - VAGUE:Velvet Assembler Graphical Front End
3297 days ago
Jitbookmark - ABACAS
3305 days ago
Surabhi Chaudharybookmark - gbtools: Interactive Visualization of Metagenome Bins in R
3267 days ago
Jitbookmark - RCircos: an R package for Circos 2D track plots
3577 days ago
Jitbookmark - CGView - Circular Genome Viewer
3455 days ago
Jitbookmark - BLAST Ring Image Generator (BRIG)
3444 days ago
Anjanabookmark
