- News
- Surabhi Chaudhary
- Update Genome Workbench 2.7.15 released
Update Genome Workbench 2.7.15 released
- Public
NCBI Genome Workbench is an integrated application for viewing and analyzing sequence data. With Genome Workbench, you can view data in publically available sequence databases at NCBI, and mix this data with your own private data.
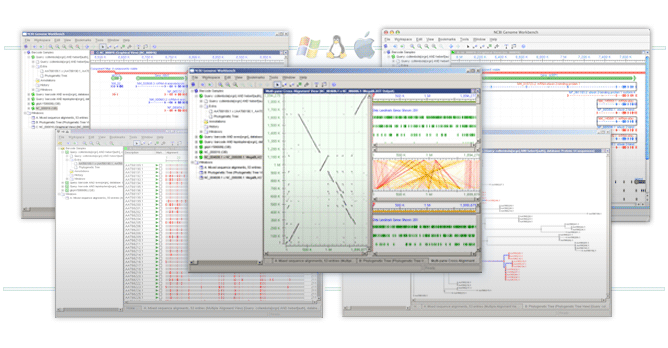
Genome Workbench can display sequence data in many ways, including graphical sequence views, various alignment views, phylogenetic tree views, and tabular views of data. It can also align your private data to data in public databases, display your data in the context of public data, and retrieve BLAST results.
Genome Workbench is built on the NCBI C++ ToolKit and uses cross-platform APIs for graphics. It runs on your local machine, and is available for Windows 2000/XP, Linux, MacOS X, and various flavors of Unix.
NCBI Genome Workbench is an integrated application for viewing and analyzing sequence data. Genome Workbench was developed entirely in-house at NCBI and makes use of the NCBI C++ ToolKit. The C++ ToolKit provides a convenient and flexible cross-platform API for managing system internals, database connections, network sockets, and the NCBI data model. In addition, the C++ ToolKit provides the Object Manager, which abstracts handling of sequences and sequence-related objects.
New Features in Genome Workbench 2.7.15
- Multiple Alignment View: implemented adaptive feature display when zooming in
- Active Objects Inspector replaces Selection Inspector. New View should offer an improved selection context examination. See Using Active Objects Inspector tutorial for more details.
- Binary packages for Linux OpenSUSE 13.1 are now available
Bug Fixes and Improvements in Genome Workbench 2.7.15
- Fixed major issue with OpenGL overlay/scrolling. Could cause crashes or view scrolling irregularities
- Multiple Pane View: fixed crash on loading BLAST results
- Graphical Sequence View: fixed crash on zooming in and out, related to SNP track
- Graphical Sequence View: fixed Go To Position dialog to give better diagnostics in case of a user error
- Graphical Sequence View: PDF export fixed rendering of Markers with commas in the name
- Text View / Flat File: fixed Mac OS rendering issues
- Text View / Flat File: performance optimization, extended capabilities of real-time rendering of molecules to tens of thousands
- File Import: optimization improvement to speed up load of files containing multiple project items
- File Import: remapping stage now shows accession.version and description of molecules, instead of plain GI numbers
- Mac OS: improved tooltips for toolbar buttons
- Phylogenetic Tree Builder Tool: improved diagnostics of errors
- Multiple Alignment View: optimizations to avoid main GUI freezes
- Open Dialog: removed duplicate elements in table of genomes (load Genome)
- PDF export: fixed issue with XREF table errors
- Tree View: fixed issues with showing Force Layout progress on Mac OS
- Tree View: PDF export fixed issues for showing labels of collapsed nodes
- Tree View: added an option to stop layout
- Tree View: broadcasting mechanism fixed not to accumulate selected nodes
Reference:
NCBI news
- A 3D Map of the Human Genome
4105 days ago
Jitendra Narayanvideo - deepTools
4140 days ago
Martin JonesNews - Genome Workbench 2.10.7
3539 days ago
Gudiya PalNews - Sequencing By Xpansion
3919 days ago
Jitendra NarayanNews - Download assemblies from NCBI
3221 days ago
BulbulNews - BIGRE Lab
4496 days ago
Radha AgarkarResearchLabs posts - Roth Lab
4382 days ago
Jitendra NarayanResearchLabs posts - Perl one-liner for bioinformatician !!!
4302 days ago
Abhimanyu Singhtop level page
