Our Sponsors
Bactopia: a flexible pipeline for complete analysis of bacterial genomes
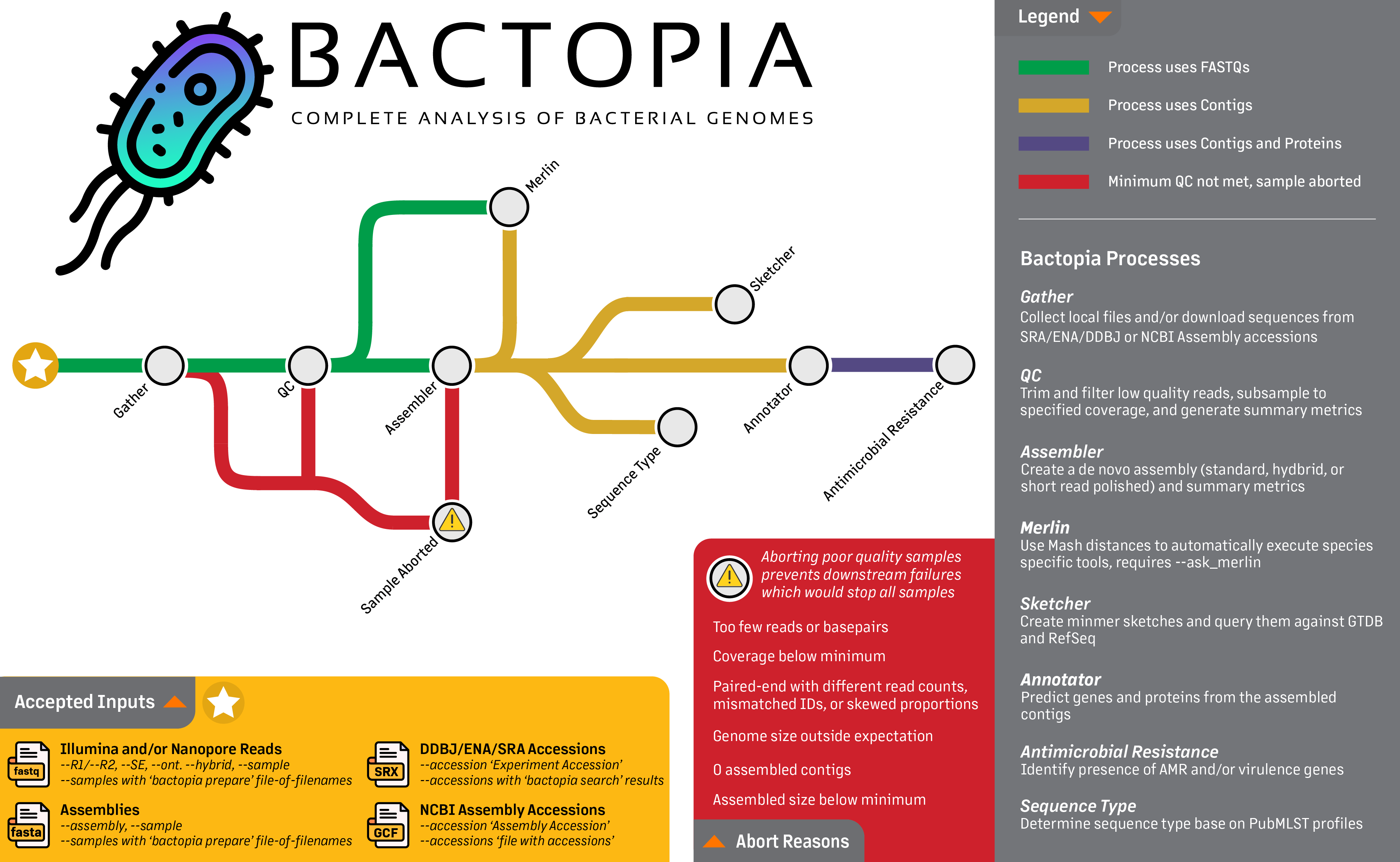
Bactopia is a flexible pipeline for complete analysis of bacterial genomes. The goal of Bactopia is process your data with a broad set of tools, so that you can get to the fun part of analyses quicker!
Bactopia was inspired by Staphopia, a workflow we (Tim Read and myself) released that is targeted towards Staphylococcus aureus genomes. Using what we learned from Staphopia and user feedback, Bactopia was developed from scratch with usability, portability, and speed in mind from the start.
Bactopia uses Nextflow to manage the workflow, allowing for support of many types of environments (e.g. cluster or cloud). Bactopia allows for the usage of many public datasets as well as your own datasets to further enhance the analysis of your sequencing. Bactopia only uses software packages available from Bioconda and Conda-Forge to make installation as simple as possible for all users.
To highlight the use of Bactopia and Bactopia Tools, we performed an analysis of 1,664 public Lactobacillus genomes, focusing on Lactobacillus crispatus, a species that is a common part of the human vaginal microbiome. The results from this analysis are published in mSystems under the title: Bactopia: a flexible pipeline for complete analysis of bacterial genomes
- Bactopia: a Flexible Pipeline for Complete Analysis of Bacterial Genomes
221 days ago
Abhibookmark - Unicycler: Hybrid assembly pipeline for bacterial genomes
2599 days ago
Jitbookmark - kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference...
2571 days ago
Jitbookmark - PGAP-X: Extension on pan-genome analysis pipeline
2525 days ago
Rahul Nayakbookmark - PyParanoid: a pipeline for rapid identification of homologous gene families in a set of genomes
1592 days ago
BioStarbookmark - NCBI Prokaryotic Genome Annotation Pipeline
2777 days ago
Jitbookmark - PhyloHerb: Phylogenomic Analysis Pipeline for Herbarium Specimens
305 days ago
LEGEbookmark - DeepVariant : an analysis pipeline that uses a deep neural network to call genetic variants from...
1793 days ago
Jitbookmark