Shikha Logwani
A bioinformatician
Our Sponsors
- News
- Shikha Logwani
- Conserved Domain Database (CDD) version 3.11 released
Conserved Domain Database (CDD) version 3.11 released
National Center for Biotechnology Information (NCBI) Conserved Domain Database (CDD) version 3.11 is now available with 596 new or updated NCBI-curated and 49,641 total domain models. The new version now contains the most recent Pfam release 27.
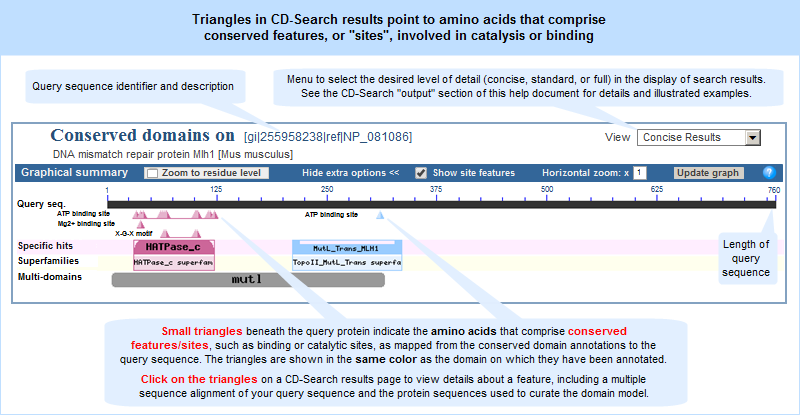
Updates to the Conserved Domain Database include:
- Position-specific score matrices (PSSMs) have been recomputed for many models in CDD, and frequency tables have been added to the PSSMs;
- The search databases distributed as part of this release can now be used with the more recent versions of RPS-BLAST (BLAST release 2.2.28 and up) using composition-based scoring. This abolishes the need to mask out compositionally biased regions in query sequences;
- Domain annotation displays in CD-Search, BATCH CD-Search, and other services now all use a uniform display style. A new display option in CD-Search and BATCH CD-Search provides “standard” results, in addition to “concise” and “full” results. “Standard” results will provide, for each region on the query sequence, the best0-scoring domain model (if any) from each of CDD’s database providers (Pfam, SMART, COG, TIGRFAMs, Protein Clusters, and the NCBI in-house curation project), but will suppress redundancy from within a single provider's results list.
You can access CDD at the Conserved Domains homepage and find updated content on the CDD FTP site.
Reference:
NCBI Website
Related items
- IgBLAST: a popular NCBI package for classifying and analyzing immunoglobulin (IG) and T cell...
2235 days ago
BioJokerNews - Biological databases !
2215 days ago
BioStarbookmark - Update Genome Workbench 2.7.15 released
4392 days ago
Surabhi ChaudharyNews - NCBI Remap
3677 days ago
Jitbookmark - MutaBind
3561 days ago
JitNews - NCBI Prokaryotic Genome Annotation Pipeline
3217 days ago
Jitbookmark - BLAST nr version 5 database, (nr_v5)
2388 days ago
JitNews - NCBI Datasets pages
969 days ago
BioStarNews
