Jit
Enthusiast computational biologist with business dream !!!
Our Sponsors
SKESA: strategic k-mer extension for scrupulous assemblies
https://github.com/ncbi/SKESA/releases
SKESA is a DeBruijn graph-based de-novo assembler designed for assembling reads of microbial genomes sequenced using Illumina. Comparison with SPAdes and MegaHit shows that SKESA produces assemblies that have high sequence quality and contiguity, handles low-level contamination in reads, is fast, and produces an identical assembly for the same input when assembled multiple times with the same or different compute resources.
Source code for SKESA is freely available at https://github.com/ncbi/SKESA/releases.
Research Paper @ Link
SKESA algorithm are as follows:
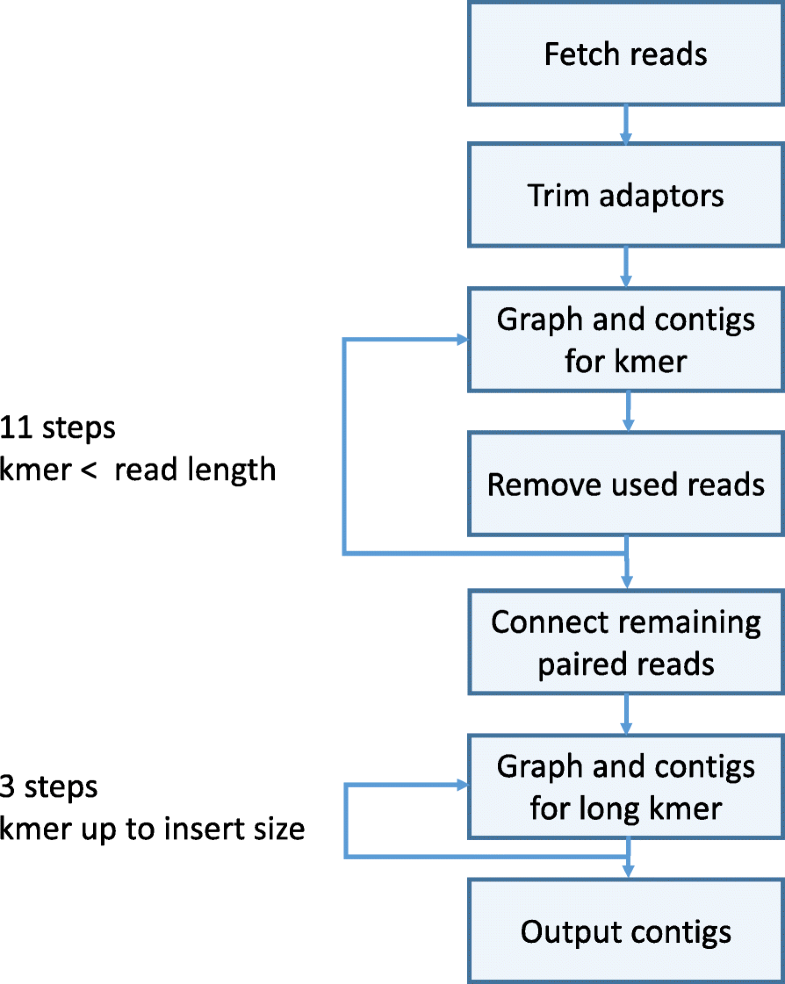
Related items
- KAD: Assessing genome assemblies using K-mer copies in assemblies and K-mer abundance in Illumina...
2091 days ago
Jitbookmark - LACHESIS: Genome Assembly with Hi-C-based Contact Probability Maps (LACHESIS)
2858 days ago
Jitbookmark - fragScaff: Genome Assembly with Contiguity Preserving Transposition
2858 days ago
Jitbookmark - COPE: an accurate k-mer-based pair-end reads connection tool to facilitate genome assembly
3017 days ago
Jitbookmark - Hagfish - assess an assembly through creative use of coverage plots
3581 days ago
Abhibookmark - KAT: a K-mer analysis toolkit to quality control NGS datasets and genome assemblies
2805 days ago
Jitbookmark - odgi: optimized dynamic genome/graph implementation
1498 days ago
Abhimanyu Singhbookmark - GMcloser: closing gaps in assemblies accurately with a likelihood-based selection of contig or...
2830 days ago
Shruti Paniwalabookmark
